The Project#
This project integrates various tools for modeling and simulating RNA structures. In addition, it includes functionalities designed to efficiently analyze molecular dynamics (MD) trajectories of labeled RNA. A key feature of the project is the simulation of Förster Resonance Energy Transfer (FRET) experiments. By utilizing the pipeline described here, RNA structures can be modeled and evaluated using single-molecule FRET (smFRET) experimental data.
The Concept#
The pipeline consists of three main modules: Modeling, Simulation, and Data Analysis.
Modeling: The modeling module allows the prediction of 2D RNA structures using RNAfold. For 3D structure modeling, the pipeline integrates the Rosetta tool for RNA 3D structure prediction. In addition, a knowledge-based approach is demonstrated through PyMOL, enabling the creation of specialized structural conformations. The pipeline also provides integration for in silico fluorescence labeling using FRETlabel.
Simulation: In this module, the generated structural models can be simulated using GROMACS. Additionally, with the FRETraj tool, polarization and color-resolved photon burst simulations can be performed, as well as the determination of Accessible Contact Volumes (ACV).
Data Analysis: To process the results from MD simulations and simulate FRET distributions, the Data Analysis module organizes the workflow, utilizing FRETraj for FRET simulations. This allows for the computation and analysis of FRET distributions based on the MD trajectories.
At the end of the workflow, the calculated FRET distributions can be compared to experimental data to evaluate and refine the structural models.
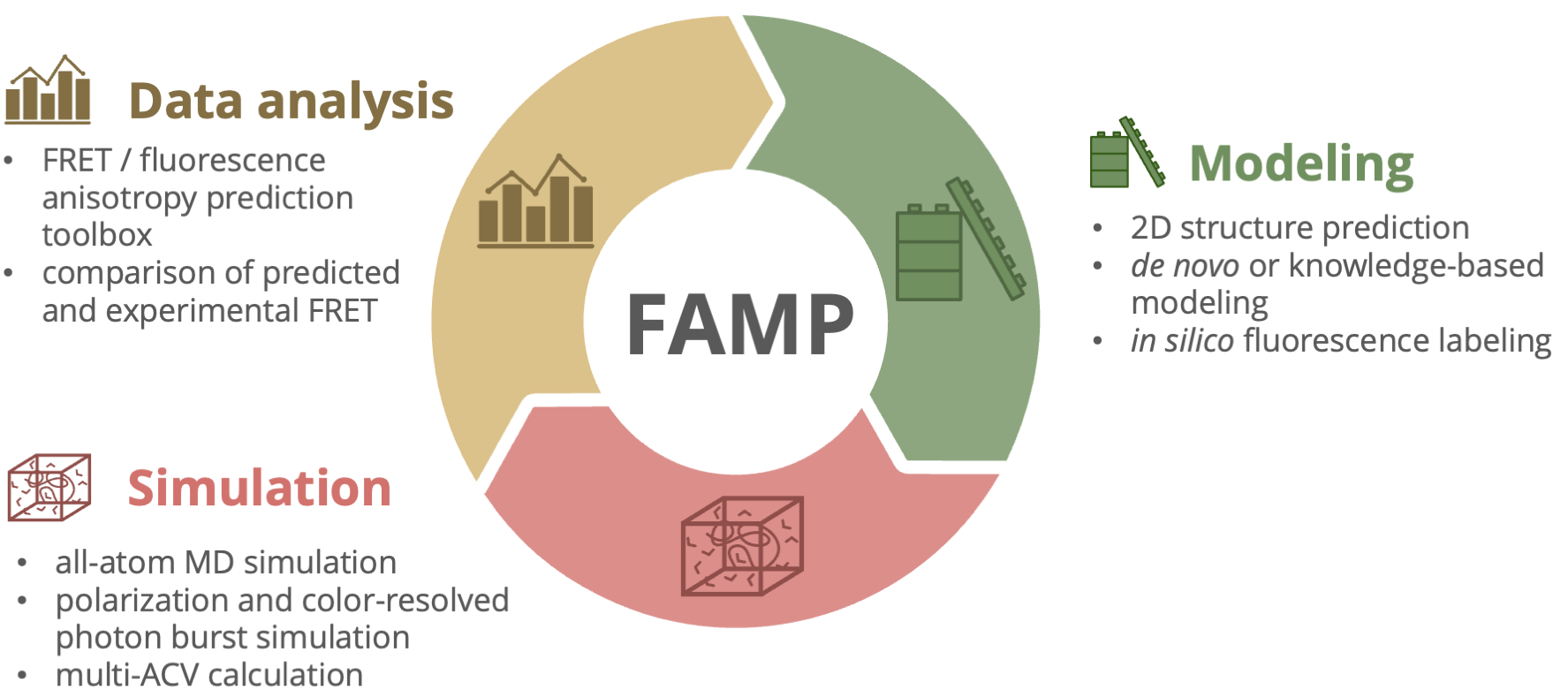
Fig. 1 Schematic overview of the FAMP pipeline. The pipeline consists of the Modeling, Simulation, and Data Analysis modules, covering 2D/3D structure prediction, molecular dynamics simulations, and FRET data analysis.#